Sharing Big Data Sets with ebulk and Wendelin

Roque Porquetto
roque (at) gmail (dot) com
https://lab.nexedi.com/u/rporchetto
This presentation aims to demonstrate how ebulk, a wrapper for embulk, can be combined with Wendelin to form an easy to use data lake capable of sharing petabytes of data grouped into data sets.
It is the result of a collaboration between Nexedi and Telecom Paristech under the "Wendelin IA" project.
Agenda
- Sharing Big Data
- Wendelin as a Data Lake
- ebulk = embulk as easy as git
- Future work
The presentation has four parts.... XXXX
Big Data Sharing
Big Data sharing is essential for research and startups. Building new A.I. models required access to large data sets. Those data sets are available to big platforms such as Google or Alibaba which tend to keep them secret. However, smaller companies or independent research teams have no access to this type of data.
Big Data sharing is also essential inside a large corporation so that production teams which produce big data sets can share it with research teams which produce A.I. models.
Specific Problems
- Huge transfer (over slow and unreliable network)
- Huge storage (with little budget)
- Many protocols (S3, HDFS, etc.)
- Many binary formats (ndarray, video, etc.)
- Trade secret
A Big Data sharing platform has two solve five problems...
XXX
XXX
XXX
XXX
Last, it should protect trade secret against the extra-territorial application of certain Laws. For exemple, any data hosted by a US platform can be accessed by NSA, no matter the location of the data centre. This is the consequence of legal obligation of US companies: the CLOUD Act. Any company willing to enforce trade secret on its data should thus stay away from US platform and use a solution with full access to source code (for auditing purpose), possibly on premise.
Existing solutions?
- Amazon S3
- Microsoft Azure
- Git Large File Storage
There are actually very few solutions to share big data sets efficiently.
Some research teams use Amazon S3. However, Amazon S3 does no provide a tool to manage a catalog of data sets, and is not suitable for data protected by trade secret (unless it is encrypted).
Most corporations tend to useMicrosoft Azure's data lake technology. However, Microsoft Azure can be fairly expensive and does not satisfy certain trade secret requirements due to the CLOUD Act.
One could also use Github as a data lake thanks to Git Large File Storage. There is even an open source implementation for Gitlab which could be combined with Sheepdog block storage and Nexedi's gitlab acceleration patch (17 times faster) to form a high performance platform.
XXX explain why those solutions are insufficient XXX
Wendelin as a Data Lake
We are now going to explain how one can easily build a Big Data sharing platform based on Wendelin framwork.
Wendelin - Out-of-core Pydata
 https://wendelin.nexedi.com/
https://wendelin.nexedi.com/
Wendelin is a big data framework designed for industrial applications based on python, NumPy, Scipy and other NumPy based libraries. It uses at its core the NEO distributed transactional NoSQL database to store petabytes of binary data ...
XXX
Data Ingestion
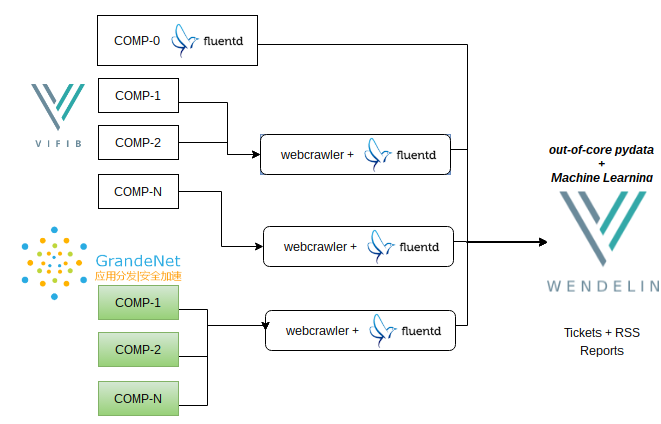
Wendelin has been implemented mainly in streaming applications where data is ingested in real time from wind turbines, using fluentd middleware by Treasure Data, a pioneer of big data platforms. Fluentd has proven to be useful thanks to its multi-protocol nature which ensure perfect interoperability and to its buffering technology which ensures no data is lost over poor networks.
Petabytes of out-of-core streams or arrays
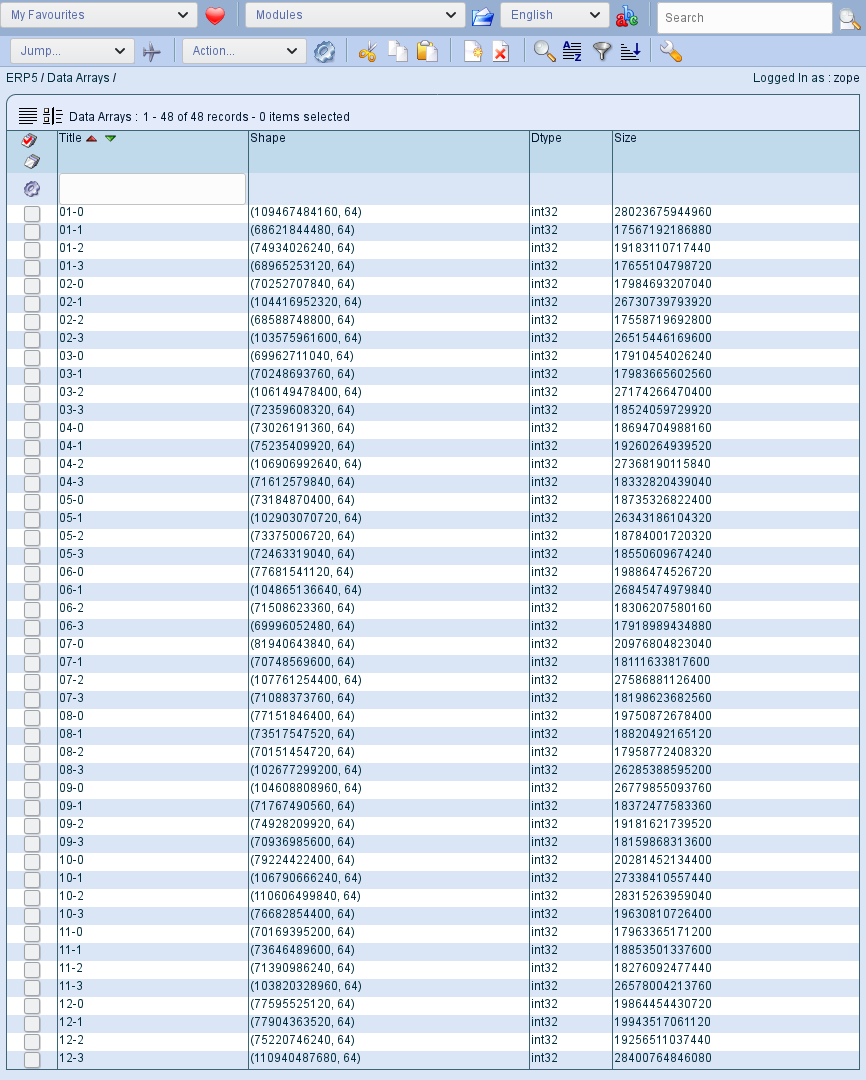
Wendelin can store petabytes of out-of-core streams or arrays. This is a very important feature because it means that any batch script on server side can process arrays or streams that are much larger than RAM. XXX explain XXX
ebulk = embulk as easy as git
Based on our success with fluentd, we selected another tool from Treasure Data: embulk. Embulk provides the same kind of interoperability. However, it was a bit complex to use.
Therefore we created a wrapper called ebulk which imitated git syntax yet is based on embulk. We use ebulk to push, pull, add, etc. big data to a data set.
Data Set UI
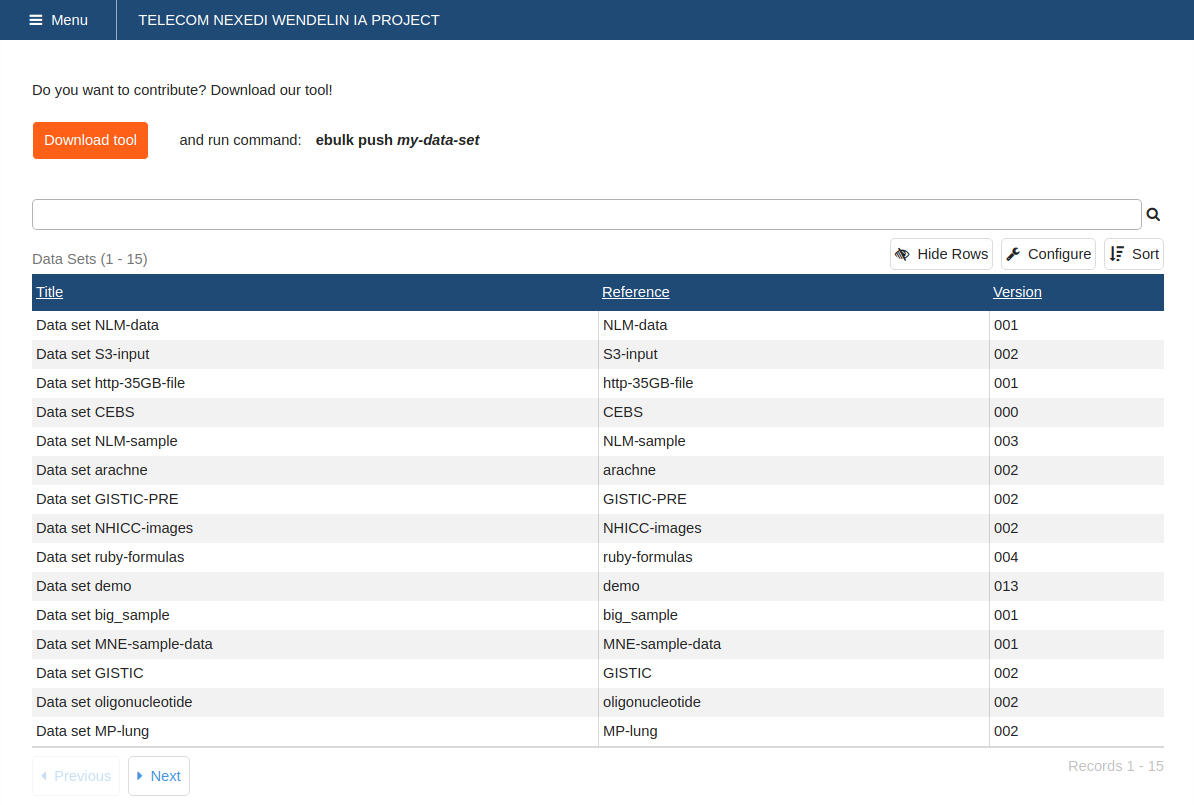
Data Set UI
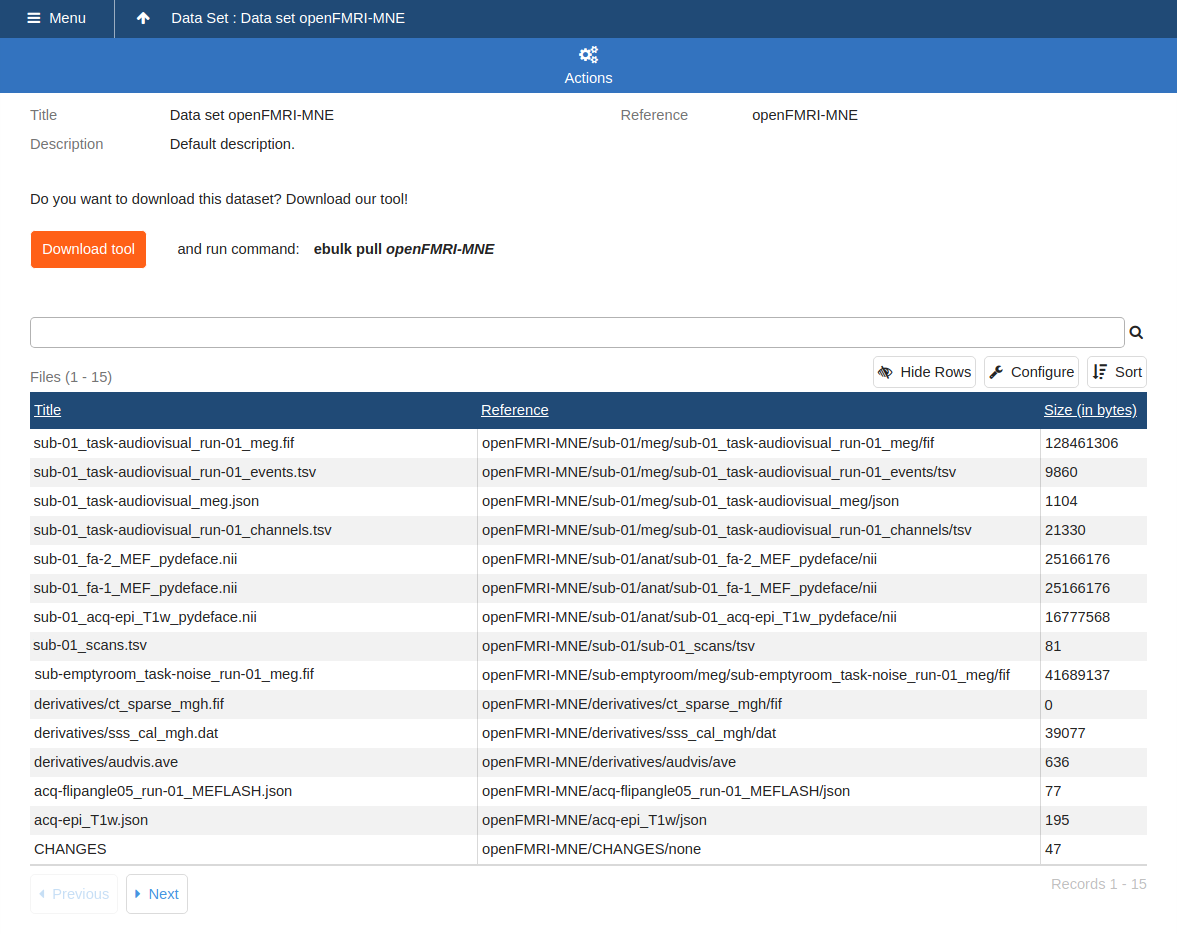
Data Set UI
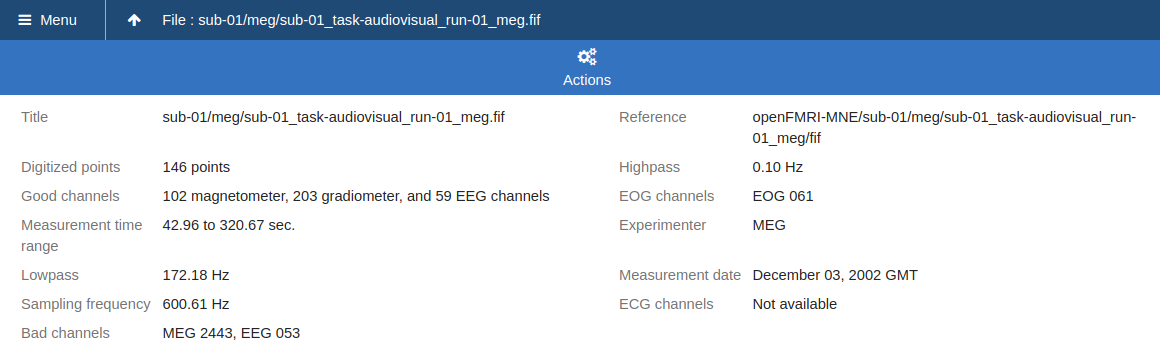
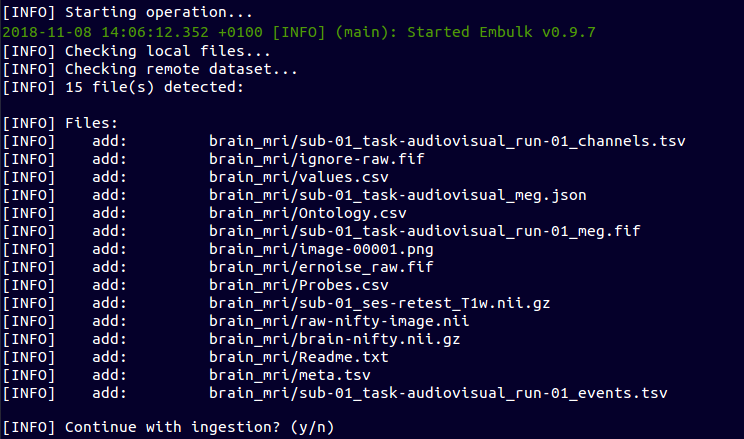
Create and Upload a Data Set
~ $ ebulk init brain_mri
~ $ ebulk push brain_mri
Create and Upload a Data Set
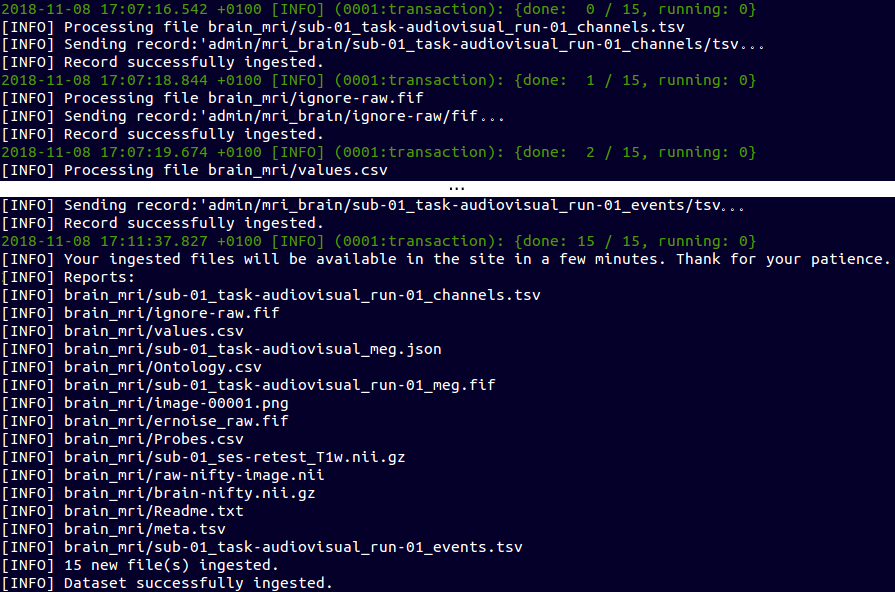
Create and Upload a Data Set
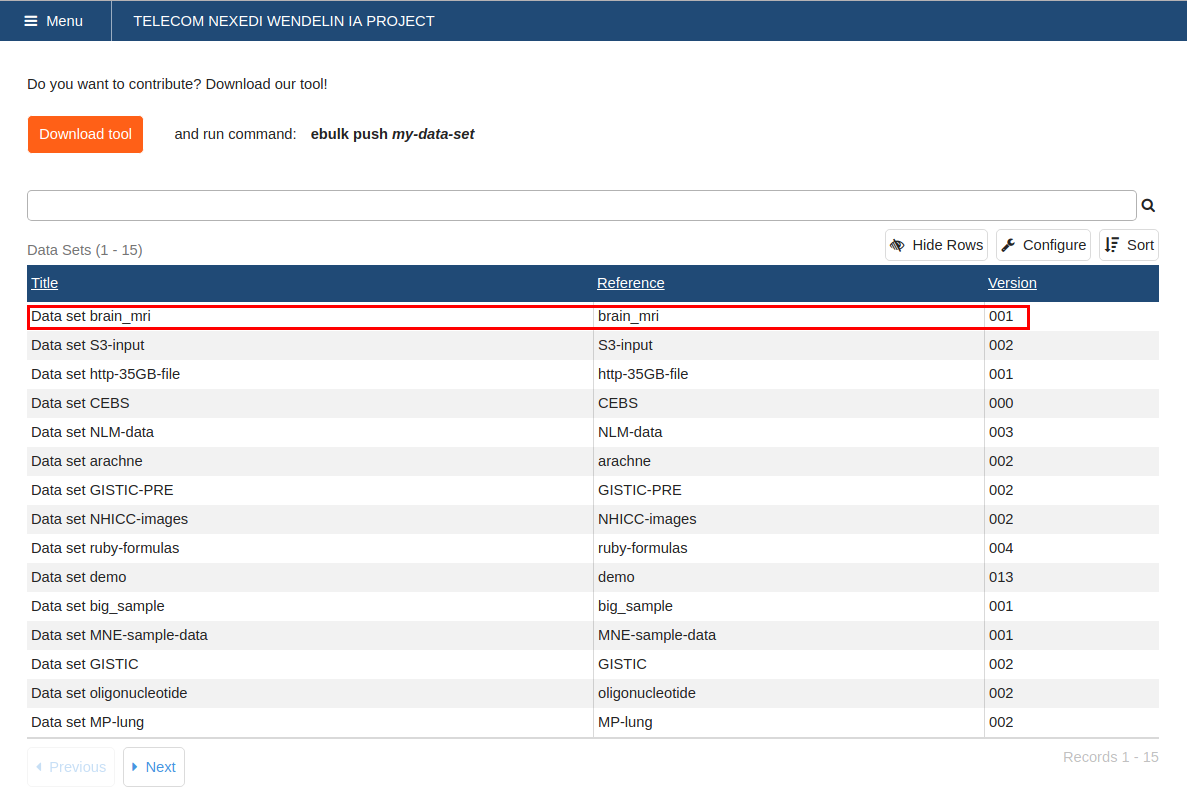
Create and Upload a Data Set
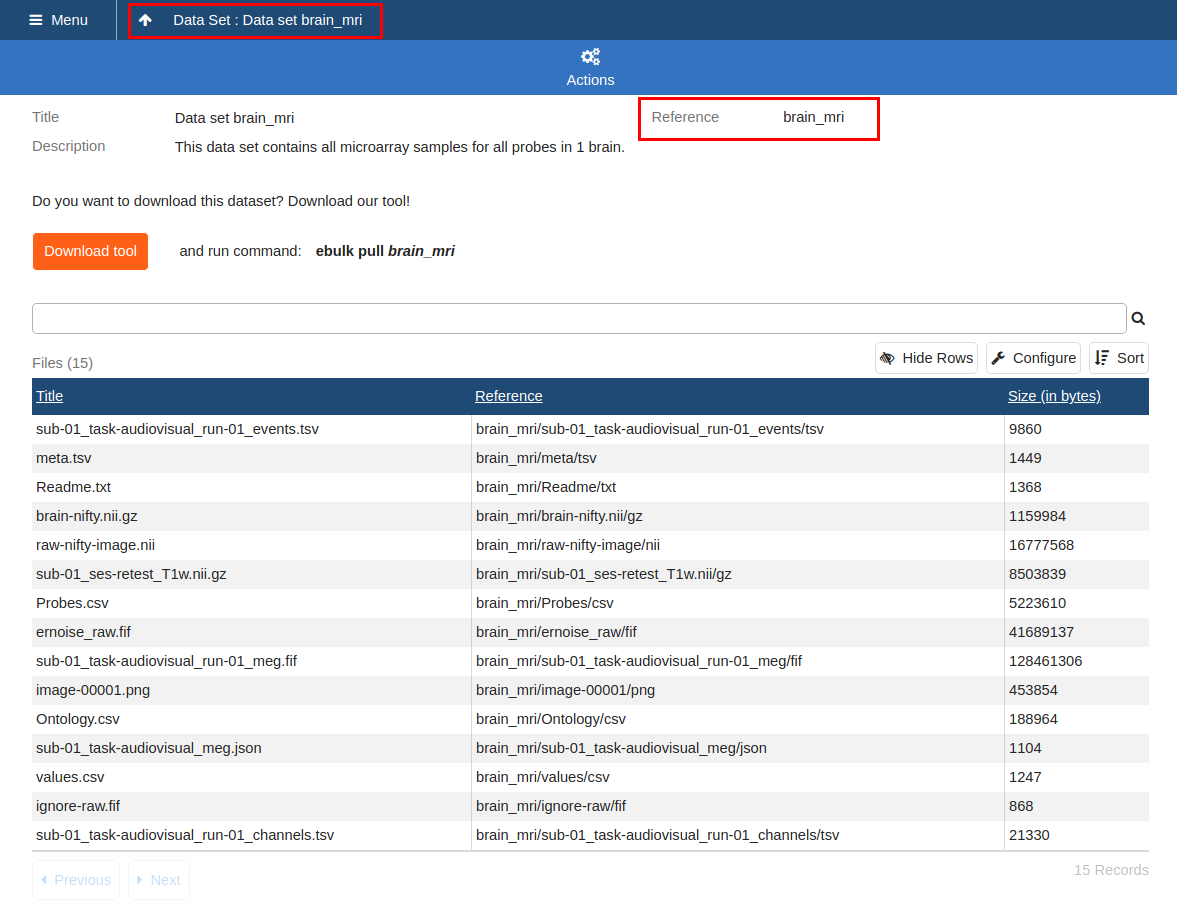
Download a Data Set
~ $ ebulk pull brain_mri
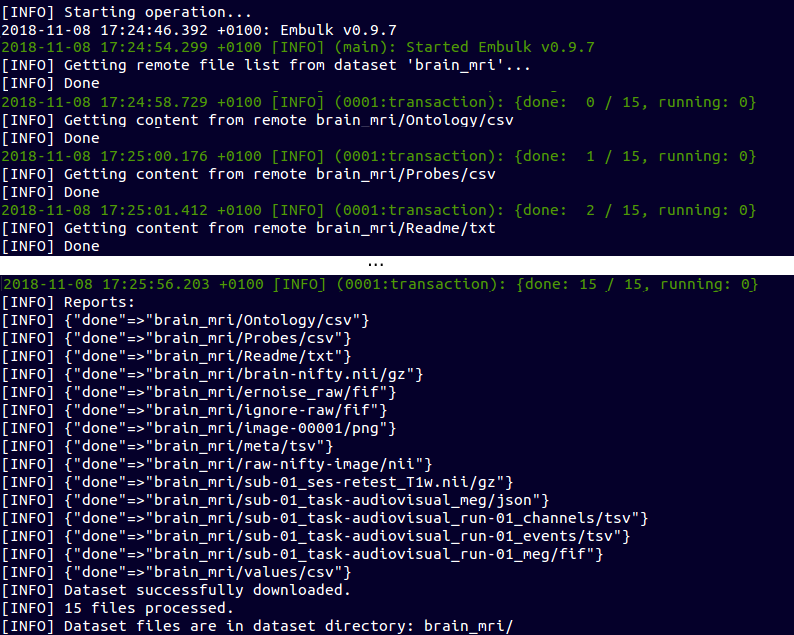
Download a Data Set
~ $ ls brain_mri_12

Contribute to a Data Set
~ $ cp ~/new_mri/2038273.mri brain_mri
~ $ mv brain_mri/Probes.csv brain_mri/initial_probes.csv
~ $ vi brain_mri/Readme.txt
~ $ rm brain_mri/meta.tsv
~ $ ebulk status brain_mri
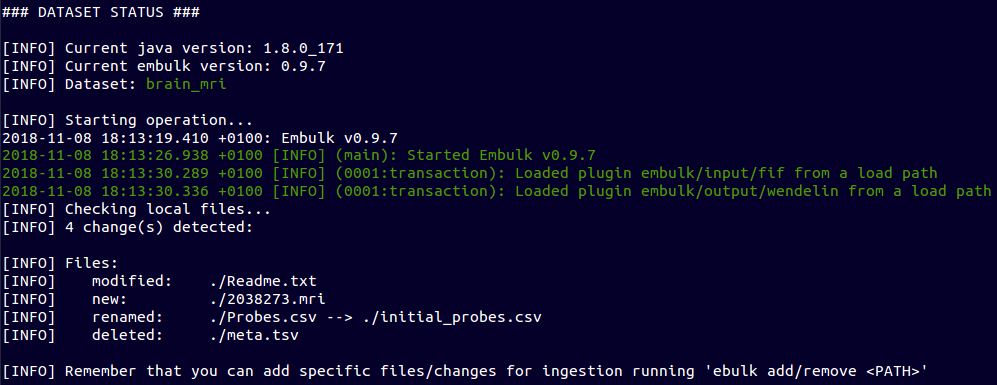
Contribute to a Data Set
~ $ ebulk add brain_mri/2038273.mri
~ $ ebulk add brain_mri/initial_probes.csv
~ $ ebulk add brain_mri/Readme.txt
~ $ ebulk remove brain_mri/meta.tsv
~ $ ebulk push brain_mri
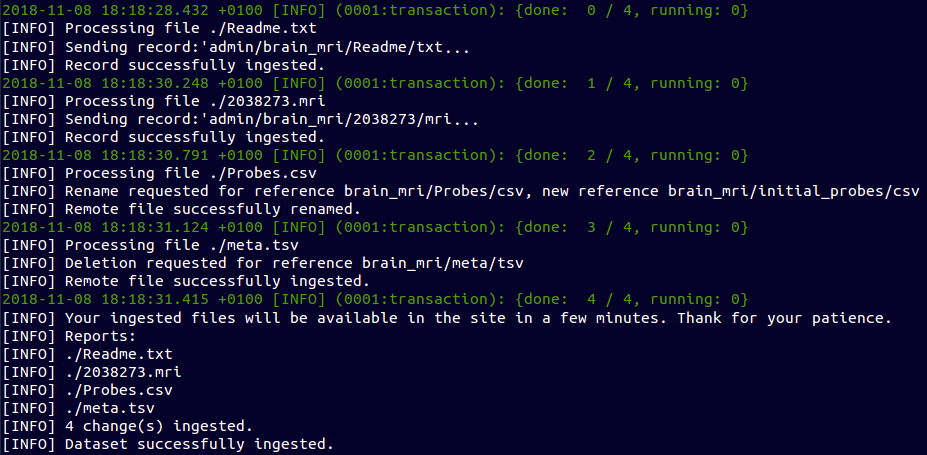
Contribute to a Data Set - Partial ingestion
~ $ ebulk push brain_mri --directory new_mri/
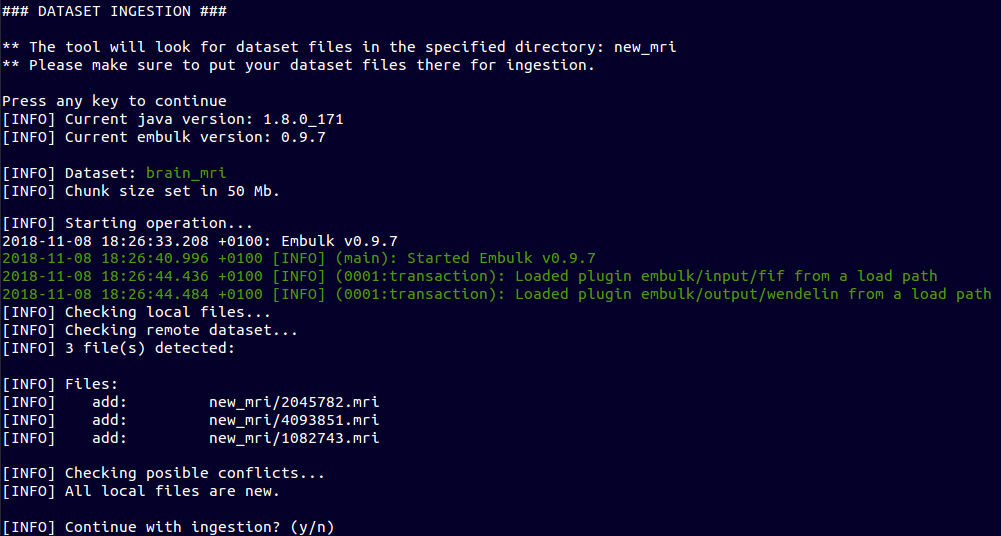
Resume interrupted operations
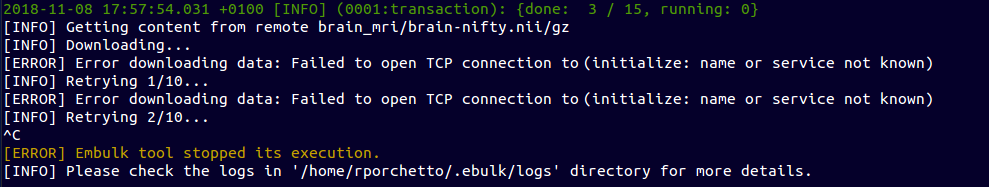
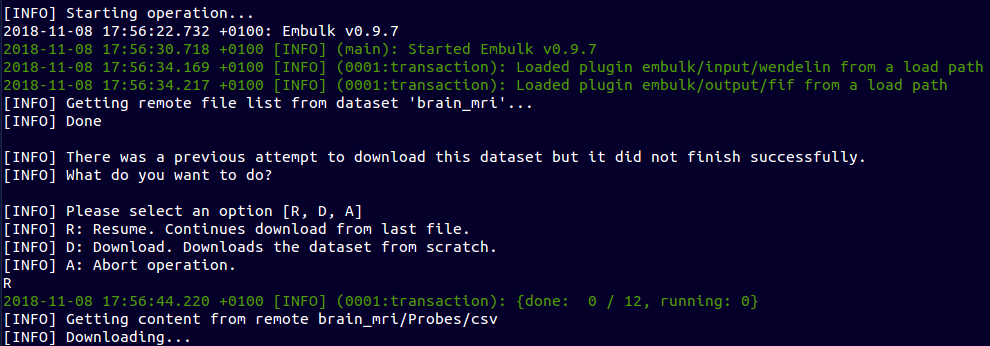
~ $ ebulk pull brain_mri
Push from different protocols
- FTP transfer
- HTTP request to URL
- Amazon S3
- Extendable to wrap and use other existing Embulk plugins like HDFS, GCS, etc
~ $ ebulk push brain_mri --storage <protocol>
Push from Amazon S3
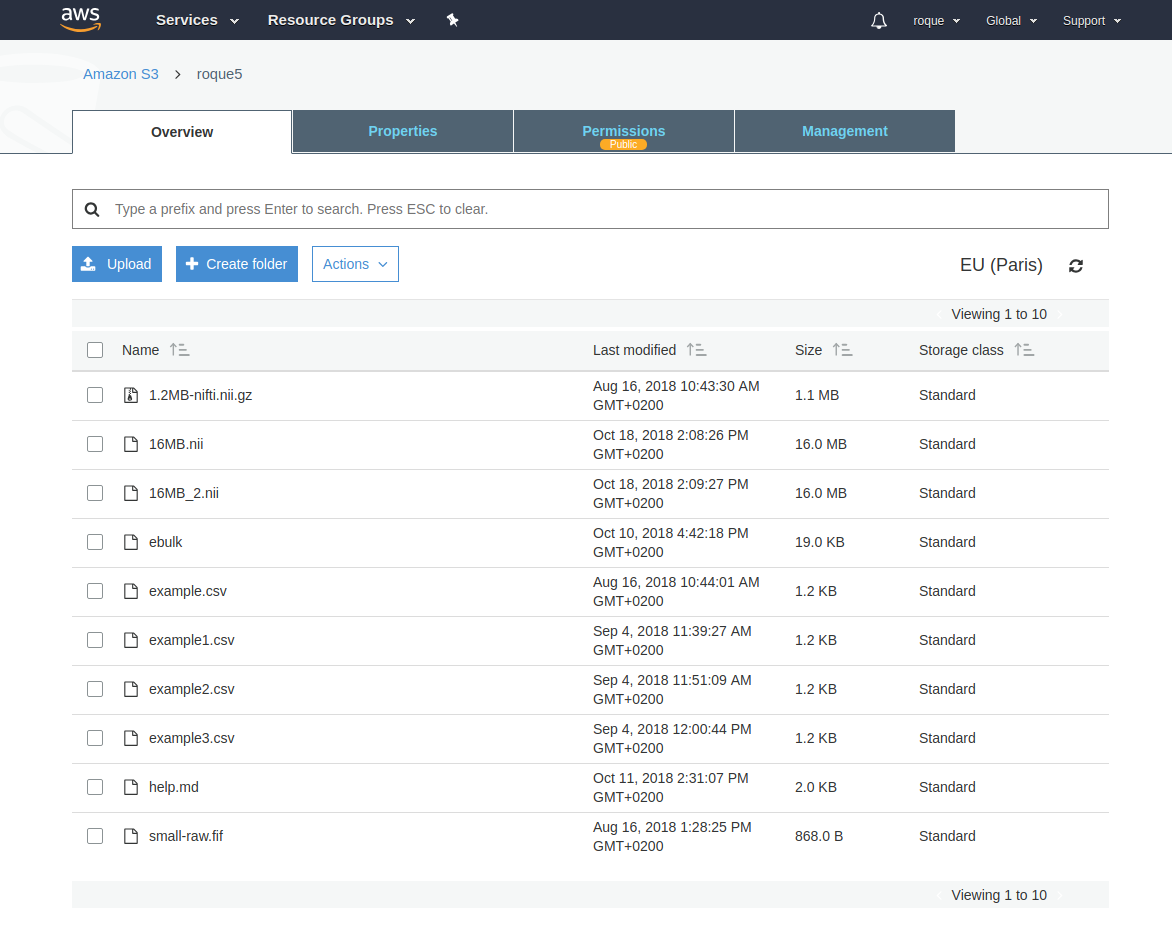
Push from Amazon S3
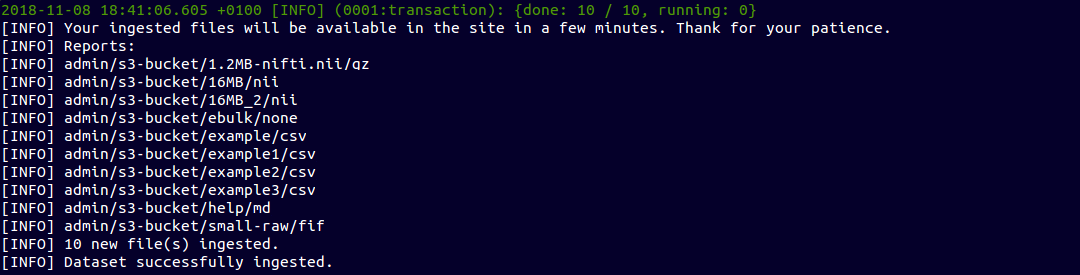
~ $ ebulk push S3-input --storage s3
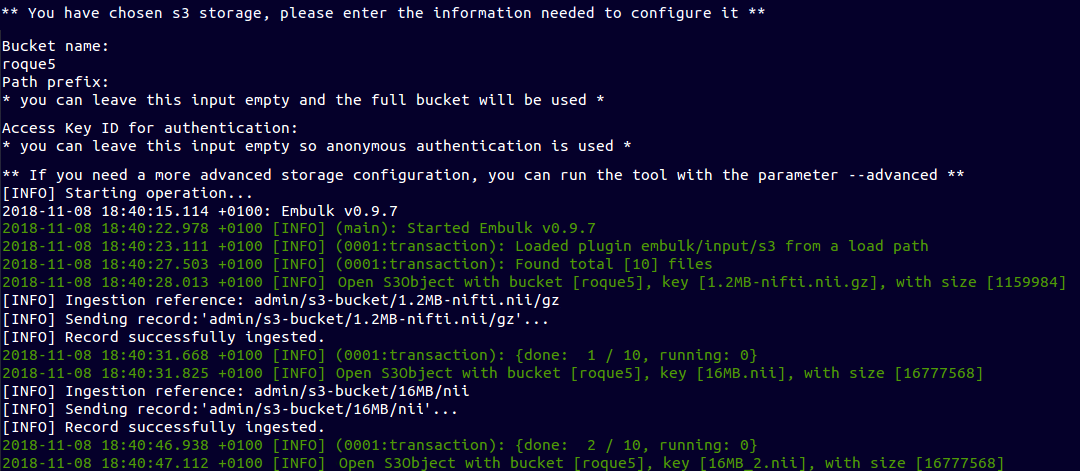
Push from Amazon S3
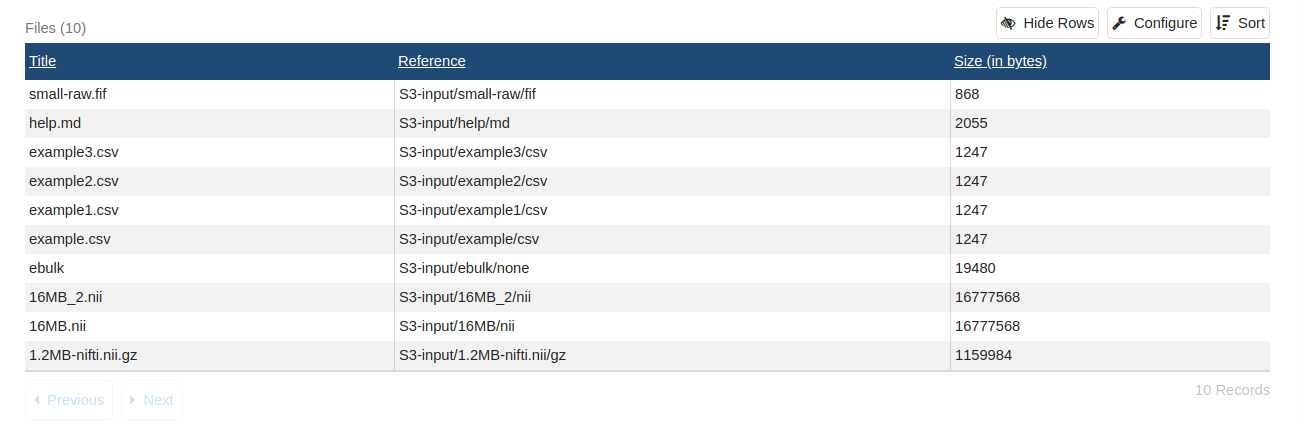
ebulk summary
- Upload
- Download
- Check local changes and stage for upload
- Contribute
- Resume operations
- Upload using different protocols
ebulk summary
ebulk push
ebulk pull
ebulk add/remove
ebulk status
ebulk push --storage
Thank You
- Nexedi GmbH
- 147 Rue du Ballon
- 59110 La Madeleine
- France